Inspired by these beautiful examples, I have tried to use the ggtern R package to draw a few ternary maps showing how the H mag of various asteroid families varies as a function of q, i.e. the perihelium distance.
As usual, the starting point ...
....consisted in downloading the parameters a,e,H from the JPL Small-Body Database Search Engine.
....consisted in downloading the parameters a,e,H from the JPL Small-Body Database Search Engine.
After that, I divided the H range into three parts: Hhigh (upper 1/3 of the H range), Hmed (medium 1/3 of the H range), Hlow (lower 1/3 of the H range).
Then, I determined the perihelium percentiles (and their median q_median).
Finally, I built a table displaying the proportions of Hhigh, Hmed, Hlow in the various perihelium percentiles (of course, the sum of the proportions in every percentile has to be 1 while the three individual proportions might be different from 1/3 - thus, a ternary map can be used to represent the various proportions).
If you are interested in the R programming details, look at the bottom of this post: I have used the R markdown language to embed the R script used to draw the ternary map for Apollo asteroids (by changing the name of Apollo with Amor, Imb, Omb, Trojan, Mars Crossers, TNO ... I got the similar maps for these other asteroid families).
Kind Regards,
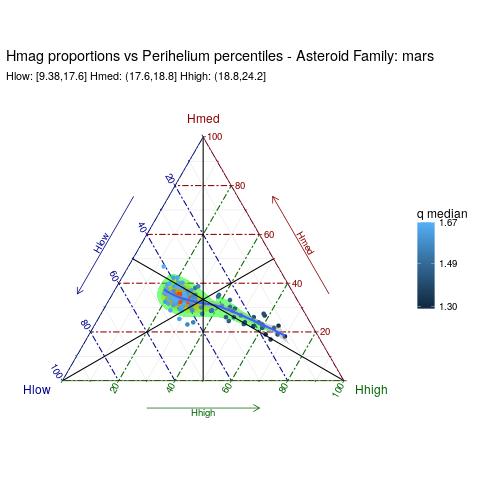
How to read the map:
- every perihelium percentile is represented by a dot
- the dot colour is associated to the median value of the perihelium percentile
- every dot has three coordinates (the sum has to be 1) representing the proportions of Hhigh, Hmed and Hlow asteroids.
- a density area is also displayed to highlight where the dots are denser
- a loess line is also drawn, though not so much significant, to give an idea of the macroscopic overall average behaviour
Note that, by definition:
- a dot near the barycenter has the same proportions of Hhigh, Hmed and Hlow (1/3+1/3+1/3).
- a dot very near to a vertex (if any) - say Hlow, it would represent a percentile where Hlow is almost equal to 1, while Hmed and High are negligible.
- a dot on a median - say the one connecting Hlow, it would represent a percentile where the High and Hmed proportions are the same.
Amor
IMB
OMB
Trojan
TNO
Appendix - R script
#citation
citation();citation("reshape2");citation("ggtern");citation("ggplot2")##
## To cite R in publications use:
##
## R Core Team (2016). R: A language and environment for
## statistical computing. R Foundation for Statistical Computing,
## Vienna, Austria. URL https://www.R-project.org/.
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {R: A Language and Environment for Statistical Computing},
## author = {{R Core Team}},
## organization = {R Foundation for Statistical Computing},
## address = {Vienna, Austria},
## year = {2016},
## url = {https://www.R-project.org/},
## }
##
## We have invested a lot of time and effort in creating R, please
## cite it when using it for data analysis. See also
## 'citation("pkgname")' for citing R packages.##
## To cite reshape2 in publications use:
##
## Hadley Wickham (2007). Reshaping Data with the reshape Package.
## Journal of Statistical Software, 21(12), 1-20. URL
## http://www.jstatsoft.org/v21/i12/.
##
## A BibTeX entry for LaTeX users is
##
## @Article{,
## title = {Reshaping Data with the {reshape} Package},
## author = {Hadley Wickham},
## journal = {Journal of Statistical Software},
## year = {2007},
## volume = {21},
## number = {12},
## pages = {1--20},
## url = {http://www.jstatsoft.org/v21/i12/},
## }##
## To cite package 'ggtern' in publications use:
##
## Nicholas Hamilton (2016). ggtern: An Extension to 'ggplot2', for
## the Creation of Ternary Diagrams. R package version 2.2.0.
## https://CRAN.R-project.org/package=ggtern
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {ggtern: An Extension to 'ggplot2', for the Creation of Ternary Diagrams},
## author = {Nicholas Hamilton},
## year = {2016},
## note = {R package version 2.2.0},
## url = {https://CRAN.R-project.org/package=ggtern},
## }##
## To cite ggplot2 in publications, please use:
##
## H. Wickham. ggplot2: Elegant Graphics for Data Analysis.
## Springer-Verlag New York, 2009.
##
## A BibTeX entry for LaTeX users is
##
## @Book{,
## author = {Hadley Wickham},
## title = {ggplot2: Elegant Graphics for Data Analysis},
## publisher = {Springer-Verlag New York},
## year = {2009},
## isbn = {978-0-387-98140-6},
## url = {http://ggplot2.org},
## }## Loading required package: ggplot2## --
## Consider donating at: http://ggtern.com
## Even small amounts (say $10-50) are very much appreciated!
## Remember to cite, run citation(package = 'ggtern') for further info.
## --##
## Attaching package: 'ggtern'## The following objects are masked from 'package:ggplot2':
##
## %+%, aes, annotate, calc_element, ggplot, ggplot_build,
## ggplot_gtable, ggplotGrob, ggsave, layer_data, theme,
## theme_bw, theme_classic, theme_dark, theme_gray, theme_light,
## theme_linedraw, theme_minimal, theme_void# file name
name="apollo"
#read file
p<-read.csv(name)
# remove rows where H is not available
p<-p[which(!is.na(p$H)),]
# calculate perihelium q
p$q<-p$a*(1-p$e)
# divide H range into three equal parts
p$HT<-cut(p$H,quantile(p$H,probs=c(0,1/3,2/3,1)),include.lowest=T)
# calculate perihelium percentiles
p$qperc<-cut(p$q,quantile(p$q,probs=seq(0,1,0.01)),include.lowest=T)
# calculate q median for every q percentile
p$qm<-tapply(p$q,list(p$qperc),median)[p$qperc]
# check
head(p[,c(1,2,8,6,9:11)]);summary(p[,c(1,2,8,6,9:11)])## a e q H HT qperc
## 1 1.078078 0.8268746 0.1866427 16.90 [12.4,20.8] [0.07066,0.2324]
## 2 1.245393 0.3353925 0.8276972 15.60 [12.4,20.8] (0.8267,0.8319]
## 3 1.367258 0.4359054 0.7712628 14.23 [12.4,20.8] (0.7681,0.7748]
## 4 1.470241 0.5599397 0.6469946 16.25 [12.4,20.8] (0.6439,0.6558]
## 5 2.259355 0.6061736 0.8897937 15.54 [12.4,20.8] (0.8885,0.893]
## 6 1.460856 0.6146128 0.5629953 14.85 [12.4,20.8] (0.5582,0.5704]
## qm
## 1 0.1835429
## 2 0.8292025
## 3 0.7709175
## 4 0.6506210
## 5 0.8904017
## 6 0.5642647## a e q H
## Min. : 1.000 Min. :0.02097 Min. :0.07066 Min. :12.40
## 1st Qu.: 1.294 1st Qu.:0.36393 1st Qu.:0.69437 1st Qu.:20.00
## Median : 1.643 Median :0.51276 Median :0.85776 Median :22.60
## Mean : 1.724 Mean :0.49672 Mean :0.80022 Mean :22.57
## 3rd Qu.: 2.090 3rd Qu.:0.62793 3rd Qu.:0.95425 3rd Qu.:25.17
## Max. :17.827 Max. :0.96910 Max. :1.01697 Max. :33.20
##
## HT qperc qm
## [12.4,20.8]:2674 [0.07066,0.2324]: 79 Min. :0.1835
## (20.8,24.4]:2621 (0.2324,0.2897] : 79 1st Qu.:0.6974
## (24.4,33.2]:2565 (0.3296,0.3655] : 79 Median :0.8574
## (0.3981,0.4277] : 79 Mean :0.8003
## (0.4277,0.4519] : 79 3rd Qu.:0.9538
## (0.4797,0.4976] : 79 Max. :1.0160
## (Other) :7386# build a table counting Hhigh, Hmed, Hlow in every q percentile
zz<-table(p$qm,p$HT)
# convert table so as to show proportions
zz<-zz/apply(zz,1,sum)
zz<-as.data.frame(zz)
colnames(zz)<-c("qm","hband","Freq")
zz<-dcast(zz,qm~hband)## Using Freq as value column: use value.var to override.colnames(zz)<-c("qm","Hlow","Hmed","Hhigh")
zz$qm<-as.numeric(as.character(zz$qm))
# check
round(head(zz),3) ## qm Hlow Hmed Hhigh
## 1 0.184 0.772 0.228 0.000
## 2 0.260 0.696 0.278 0.025
## 3 0.305 0.731 0.244 0.026
## 4 0.349 0.696 0.253 0.051
## 5 0.380 0.782 0.179 0.038
## 6 0.414 0.582 0.392 0.025# define strings
Hlow<-paste("Hlow:",names(summary(p$HT))[1])
Hmed<-paste("Hmed:",names(summary(p$HT))[2])
Hhigh<-paste("Hhigh:",names(summary(p$HT))[3])
# plot ternary map
myplot<-ggtern(zz,aes(Hlow,Hmed,Hhigh))
+stat_density_tern(geom='polygon',aes(fill=..level..),alpha=0.5)
+theme_rgbw()
+scale_fill_gradient(name="density",low="green",high="red",guide=F)
+geom_point(aes(col=qm))+geom_smooth_tern()
+scale_color_continuous(name="q median",breaks=round(seq(min(zz$qm),
max(zz$qm),length.out = 3),2),
limits=round(c(min(zz$qm),max(zz$qm)),2))
+ggtitle(paste("Hmag proportions vs Perihelium percentiles - Asteroid Family:",name),
subtitle=paste(Hlow,Hmed,Hhigh))
+geom_Risoprop(value=0.5)
+geom_Lisoprop(value = 0.5)
+geom_Tisoprop(value = 0.5)





No comments:
Post a Comment
Note: Only a member of this blog may post a comment.